gwasrapidd
gwasrapidd: an R package to query, download and wrangle GWAS Catalog data
Science Score: 13.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
○codemeta.json file
-
○.zenodo.json file
-
✓DOI references
Found 9 DOI reference(s) in README -
○Academic publication links
-
○Committers with academic emails
-
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (16.7%) to scientific vocabulary
Keywords
association-studies
gwas-catalog
human
r
rest-client
snp
trait
trait-ontology
Last synced: 6 months ago
·
JSON representation
Repository
gwasrapidd: an R package to query, download and wrangle GWAS Catalog data
Basic Info
- Host: GitHub
- Owner: ramiromagno
- License: other
- Language: R
- Default Branch: master
- Homepage: https://rmagno.eu/gwasrapidd/
- Size: 21.4 MB
Statistics
- Stars: 98
- Watchers: 2
- Forks: 14
- Open Issues: 4
- Releases: 0
Topics
association-studies
gwas-catalog
human
r
rest-client
snp
trait
trait-ontology
Created over 7 years ago
· Last pushed 9 months ago
Metadata Files
Readme
Contributing
License
Code of conduct
README.Rmd
---
output: github_document
---
```{r setup, include = FALSE}
knitr::opts_chunk$set(
collapse = TRUE,
# comment = "#>",
fig.path = "man/figures/README-",
out.width = "100%"
)
```
# gwasrapidd  [](https://CRAN.R-project.org/package=gwasrapidd)
[](https://doi.org/10.1093/bioinformatics/btz605)
[](https://www.altmetric.com/details/64505748)
[](https://github.com/ramiromagno/gwasrapidd/actions/workflows/R-CMD-check.yaml)
[](https://opensource.org/license/mit)
The goal of `{gwasrapidd}` is to provide programmatic access to the [NHGRI-EBI
Catalog](https://www.ebi.ac.uk/gwas) of published genome-wide association
studies.
Get started by reading the
[documentation](https://rmagno.eu/gwasrapidd/articles/gwasrapidd.html).
## Installation
Install `{gwasrapidd}` from CRAN:
``` r
install.packages("gwasrapidd")
```
## Cheatsheet
[](https://CRAN.R-project.org/package=gwasrapidd)
[](https://doi.org/10.1093/bioinformatics/btz605)
[](https://www.altmetric.com/details/64505748)
[](https://github.com/ramiromagno/gwasrapidd/actions/workflows/R-CMD-check.yaml)
[](https://opensource.org/license/mit)
The goal of `{gwasrapidd}` is to provide programmatic access to the [NHGRI-EBI
Catalog](https://www.ebi.ac.uk/gwas) of published genome-wide association
studies.
Get started by reading the
[documentation](https://rmagno.eu/gwasrapidd/articles/gwasrapidd.html).
## Installation
Install `{gwasrapidd}` from CRAN:
``` r
install.packages("gwasrapidd")
```
## Cheatsheet
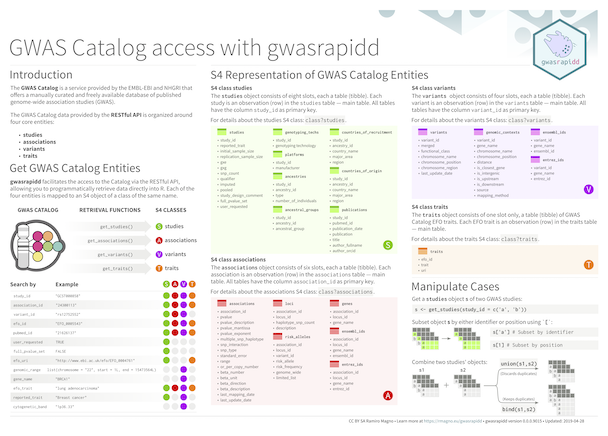 ## Example
Get studies related to triple-negative breast cancer:
```{r example1}
library(gwasrapidd)
studies <- get_studies(efo_trait = 'triple-negative breast cancer')
studies@studies[1:4]
```
Find associated variants with study `r studies@studies$study_id[1]`:
```{r example2}
variants <- get_variants(study_id = 'GCST002305')
variants@variants[c('variant_id', 'functional_class')]
```
## Citing this work
`{gwasrapidd}` was published in Bioinformatics in 2019:
https://doi.org/10.1093/bioinformatics/btz605.
To generate a citation for this publication from within R:
```{r citation}
citation('gwasrapidd')
```
## Code of Conduct
Please note that the `{gwasrapidd}` project is released with a [Contributor Code
of Conduct](https://rmagno.eu/gwasrapidd/CODE_OF_CONDUCT.html). By contributing
to this project, you agree to abide by its terms.
## Similar projects
- Bioconductor R package *gwascat* by Vincent J Carey:
[https://www.bioconductor.org/packages/release/bioc/html/gwascat.html](https://www.bioconductor.org/packages/release/bioc/html/gwascat.html)
- Web application *PhenoScanner V2* by Mihir A. Kamat, James R. Staley, and
others:
[doi.org/10.1093/bioinformatics/btz469](https://doi.org/10.1093/bioinformatics/btz469)
- Web application *GWEHS: Genome-Wide Effect sizes and Heritability Screener* by
Eugenio López-Cortegano and Armando Caballero:
[http://gwehs.uvigo.es/](http://gwehs.uvigo.es/)
## Acknowledgements
This work would have not been possible without the precious help from the GWAS
Catalog team, particularly [Daniel
Suveges](https://www.ebi.ac.uk/about/people/daniel-suveges).
## Example
Get studies related to triple-negative breast cancer:
```{r example1}
library(gwasrapidd)
studies <- get_studies(efo_trait = 'triple-negative breast cancer')
studies@studies[1:4]
```
Find associated variants with study `r studies@studies$study_id[1]`:
```{r example2}
variants <- get_variants(study_id = 'GCST002305')
variants@variants[c('variant_id', 'functional_class')]
```
## Citing this work
`{gwasrapidd}` was published in Bioinformatics in 2019:
https://doi.org/10.1093/bioinformatics/btz605.
To generate a citation for this publication from within R:
```{r citation}
citation('gwasrapidd')
```
## Code of Conduct
Please note that the `{gwasrapidd}` project is released with a [Contributor Code
of Conduct](https://rmagno.eu/gwasrapidd/CODE_OF_CONDUCT.html). By contributing
to this project, you agree to abide by its terms.
## Similar projects
- Bioconductor R package *gwascat* by Vincent J Carey:
[https://www.bioconductor.org/packages/release/bioc/html/gwascat.html](https://www.bioconductor.org/packages/release/bioc/html/gwascat.html)
- Web application *PhenoScanner V2* by Mihir A. Kamat, James R. Staley, and
others:
[doi.org/10.1093/bioinformatics/btz469](https://doi.org/10.1093/bioinformatics/btz469)
- Web application *GWEHS: Genome-Wide Effect sizes and Heritability Screener* by
Eugenio López-Cortegano and Armando Caballero:
[http://gwehs.uvigo.es/](http://gwehs.uvigo.es/)
## Acknowledgements
This work would have not been possible without the precious help from the GWAS
Catalog team, particularly [Daniel
Suveges](https://www.ebi.ac.uk/about/people/daniel-suveges).
Owner
- Name: Ramiro Magno
- Login: ramiromagno
- Kind: user
- Company: Pattern Institute
- Repositories: 65
- Profile: https://github.com/ramiromagno
Data Scientist
GitHub Events
Total
- Issues event: 8
- Watch event: 7
- Issue comment event: 12
- Push event: 9
Last Year
- Issues event: 8
- Watch event: 7
- Issue comment event: 12
- Push event: 9
Committers
Last synced: 11 months ago
Top Committers
| Name | Commits | |
|---|---|---|
| Ramiro Magno | r****o@g****m | 373 |
| Pradeep Eranti | e****9@g****m | 1 |
| Katrin Leinweber | k****i@p****e | 1 |
| Hadley Wickham | h****m@g****m | 1 |
Committer Domains (Top 20 + Academic)
posteo.de: 1
Issues and Pull Requests
Last synced: 6 months ago
All Time
- Total issues: 50
- Total pull requests: 4
- Average time to close issues: about 1 month
- Average time to close pull requests: 19 days
- Total issue authors: 26
- Total pull request authors: 3
- Average comments per issue: 3.38
- Average comments per pull request: 1.25
- Merged pull requests: 3
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 6
- Pull requests: 0
- Average time to close issues: about 2 months
- Average time to close pull requests: N/A
- Issue authors: 5
- Pull request authors: 0
- Average comments per issue: 1.5
- Average comments per pull request: 0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- ramiromagno (15)
- mightyphil2000 (6)
- MattCloward (3)
- Shicheng-Guo (2)
- jyjo43043ver1 (2)
- peranti (2)
- Dicky84 (1)
- z1242099843 (1)
- tzup (1)
- milcs40 (1)
- jinjinzhou (1)
- lizlitkowski (1)
- totajuliusd (1)
- abhiachoudhary (1)
- huyou0916 (1)
Pull Request Authors
- peranti (2)
- katrinleinweber (1)
- hadley (1)
Top Labels
Issue Labels
bug (10)
question (4)
enhancement (4)
documentation (3)
help wanted (1)
Pull Request Labels
documentation (2)
Packages
- Total packages: 1
-
Total downloads:
- cran 1,014 last-month
- Total dependent packages: 0
- Total dependent repositories: 3
- Total versions: 6
- Total maintainers: 1
cran.r-project.org: gwasrapidd
'REST' 'API' Client for the 'NHGRI'-'EBI' 'GWAS' Catalog
- Homepage: https://github.com/ramiromagno/gwasrapidd
- Documentation: http://cran.r-project.org/web/packages/gwasrapidd/gwasrapidd.pdf
- License: MIT + file LICENSE
-
Latest release: 0.99.18
published 9 months ago
Rankings
Stargazers count: 4.8%
Forks count: 5.5%
Average: 15.3%
Dependent repos count: 16.4%
Downloads: 21.1%
Dependent packages count: 28.6%
Maintainers (1)
Last synced:
6 months ago
Dependencies
DESCRIPTION
cran
- R >= 3.2.3 depends
- assertthat * imports
- concatenate * imports
- dplyr * imports
- glue * imports
- httr * imports
- jsonlite * imports
- lubridate * imports
- magrittr * imports
- methods * imports
- pingr * imports
- plyr * imports
- progress * imports
- purrr * imports
- rlang * imports
- stringr * imports
- testthat * imports
- tibble * imports
- tidyr > 0.8.99 imports
- urltools * imports
- utils * imports
- covr * suggests
- httptest * suggests
- knitr * suggests
- rmarkdown * suggests
- spelling * suggests
.github/workflows/check-standard.yaml
actions
- actions/cache v2 composite
- actions/checkout v2 composite
- actions/upload-artifact main composite
- r-lib/actions/setup-pandoc v1 composite
- r-lib/actions/setup-r v1 composite
.github/workflows/pkgdown.yaml
actions
- JamesIves/github-pages-deploy-action v4.4.1 composite
- actions/checkout v3 composite
- r-lib/actions/setup-pandoc v2 composite
- r-lib/actions/setup-r v2 composite
- r-lib/actions/setup-r-dependencies v2 composite
.github/workflows/test-coverage.yaml
actions
- actions/cache v2 composite
- actions/checkout v2 composite
- r-lib/actions/setup-pandoc v1 composite
- r-lib/actions/setup-r v1 composite