nat
NeuroAnatomy Toolbox: An R package for the (3D) visualisation and analysis of biological image data, especially tracings of single neurons.
Science Score: 59.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
✓codemeta.json file
Found codemeta.json file -
✓.zenodo.json file
Found .zenodo.json file -
✓DOI references
Found 18 DOI reference(s) in README -
✓Academic publication links
Links to: nature.com -
✓Committers with academic emails
3 of 12 committers (25.0%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (16.6%) to scientific vocabulary
Keywords
Repository
NeuroAnatomy Toolbox: An R package for the (3D) visualisation and analysis of biological image data, especially tracings of single neurons.
Basic Info
- Host: GitHub
- Owner: natverse
- Language: R
- Default Branch: master
- Homepage: https://natverse.org/nat/
- Size: 61.2 MB
Statistics
- Stars: 69
- Watchers: 17
- Forks: 27
- Open Issues: 60
- Releases: 26
Topics
Metadata Files
README.md
nat: NeuroAnatomy Toolbox

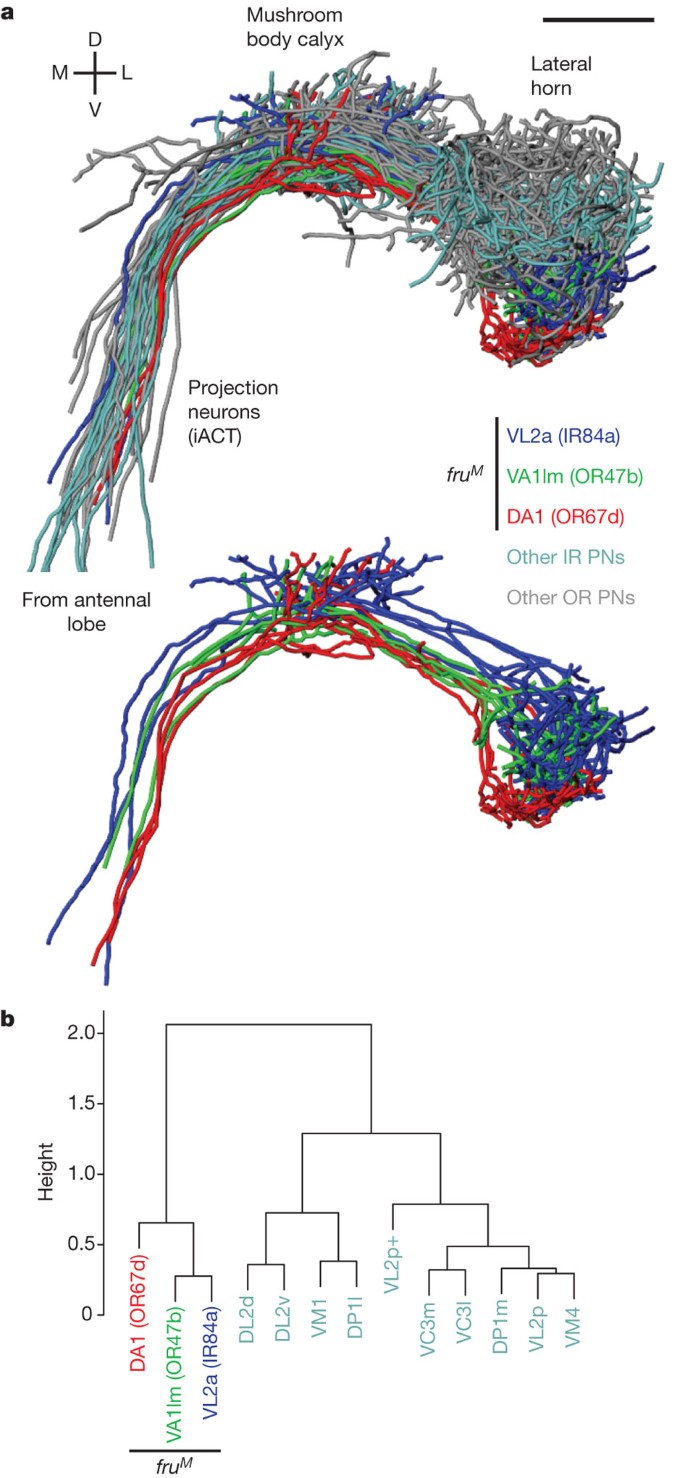
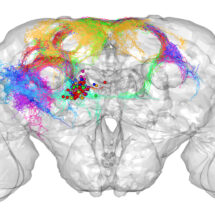
An R package for the (3D) visualisation and analysis of biological image data, especially tracings of single neurons. nat is the core package of a wider suite of neuroanatomy tools introduced at http://natverse.github.io. nat (and its ancestors) have been used in a number of papers from our group including:
Quick Start
For the impatient ...
```r
install
install.packages("nat")
use
library(nat)
plot some test data (?kcs20 for details)
Drosophila Kenyon cells processed from raw data formerly available at
flycircuit.tw
head(kcs20) open3d() plot3d(kcs20, col=type)
get help
?nat ```
Installation
A confirmed stable version of nat can be installed from CRAN.
r
install.packages("nat")
`
However, nat remains under quite active development, so if you will be using nat extensively, we generally recommend installing the latest development version directly from github using the devtools package.
```r
install devtools if required
if (!requireNamespace("devtools")) install.packages("devtools")
then install nat
devtools::install_github("natverse/nat")
```
Learn
To get an idea of what nat can do:
- Skim through the Articles listed at https://natverse.org//nat/
- Take a look at the R Markdown reports used to generate the figures for our NBLAST paper.
When you're ready to learn more:
- Read the overview package documentation
(
?natin R) - Read the Introduction to neurons article
- Check out the thematically organised function reference documentation.
- Most help pages include examples.
- Try out sample code:
- nat.examples has detailed examples for data sets from a range of model organisms and techniques
- frulhns analysis of sexually dimorphic circuits
- NBLAST figures
Help
If you want some help using nat:
- For installation issues, see the Installation vignette
- Contact nat-user Google group - we normally respond promptly and you will also be helping future users.
If you think that you have found a bug:
- Install the development version of nat using devtools (see above) and see if that helps.
- Check the github issues and
- file a bug report if this seems to be a new problem
- comment on an existing bug report
Thanks for your interest in nat!
Owner
- Name: natverse
- Login: natverse
- Kind: organization
- Website: https://natverse.org
- Repositories: 32
- Profile: https://github.com/natverse
R tools for quantitative neuroanatomy
GitHub Events
Total
- Watch event: 3
- Push event: 7
- Pull request event: 1
- Create event: 3
Last Year
- Watch event: 3
- Push event: 7
- Pull request event: 1
- Create event: 3
Committers
Last synced: over 2 years ago
Top Committers
| Name | Commits | |
|---|---|---|
| Gregory Jefferis | j****s@g****m | 2,216 |
| James Manton | a****n@g****m | 116 |
| Sri | j****n@g****m | 98 |
| Dominik Krzeminski | d****3@c****k | 57 |
| alexanderbates | a****s@g****m | 27 |
| dokato | r****2@g****m | 4 |
| Marta Costa | m****6@c****k | 3 |
| istvantaisz | i****z@g****m | 1 |
| zhihaozheng | z****n@g****m | 1 |
| Jonathan Marty | j****y@g****m | 1 |
| Ben Sutcliffe | s****f@m****k | 1 |
| Duncan Murdoch | m****n@g****m | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: over 1 year ago
All Time
- Total issues: 46
- Total pull requests: 60
- Average time to close issues: 3 months
- Average time to close pull requests: 30 days
- Total issue authors: 12
- Total pull request authors: 7
- Average comments per issue: 1.83
- Average comments per pull request: 2.7
- Merged pull requests: 46
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 2
- Pull requests: 4
- Average time to close issues: about 2 hours
- Average time to close pull requests: 4 days
- Issue authors: 2
- Pull request authors: 2
- Average comments per issue: 11.0
- Average comments per pull request: 1.0
- Merged pull requests: 2
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- jefferis (20)
- dokato (11)
- SridharJagannathan (4)
- Jsalas424 (3)
- kjheinz (1)
- szhorvat (1)
- PostPreAndCleft (1)
- artxz (1)
- dempseyg447 (1)
- mmc46 (1)
- alexanderbates (1)
Pull Request Authors
- jefferis (32)
- dokato (21)
- SridharJagannathan (4)
- alexanderbates (3)
- jonmarty (1)
- schlegelp (1)
- dmurdoch (1)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 2
-
Total downloads:
- cran 443 last-month
- Total docker downloads: 74
-
Total dependent packages: 5
(may contain duplicates) -
Total dependent repositories: 27
(may contain duplicates) - Total versions: 78
- Total maintainers: 1
proxy.golang.org: github.com/natverse/nat
- Documentation: https://pkg.go.dev/github.com/natverse/nat#section-documentation
-
Latest release: v1.10.4
published about 4 years ago
Rankings
cran.r-project.org: nat
NeuroAnatomy Toolbox for Analysis of 3D Image Data
- Homepage: https://github.com/natverse/nat
- Documentation: http://cran.r-project.org/web/packages/nat/nat.pdf
- License: GPL-3
-
Latest release: 1.8.25
published 6 months ago
Rankings
Maintainers (1)
Dependencies
- R >= 2.15.1 depends
- rgl >= 0.98.1 depends
- natcpp * enhances
- checkmate * imports
- digest * imports
- filehash >= 2.3 imports
- igraph >= 0.7.1 imports
- methods * imports
- nabor * imports
- nat.utils >= 0.4.2 imports
- pbapply * imports
- plyr * imports
- progress * imports
- stringr * imports
- yaml * imports
- zip * imports
- MASS * suggests
- Morpho * suggests
- Rvcg >= 0.17 suggests
- XML * suggests
- alphashape3d * suggests
- brotli * suggests
- httr * suggests
- jsonlite * suggests
- knitr * suggests
- plotly * suggests
- qs * suggests
- readobj * suggests
- rmarkdown * suggests
- spelling * suggests
- testthat * suggests
- actions/cache v1 composite
- actions/checkout v2 composite
- actions/upload-artifact v2 composite
- r-lib/actions/setup-pandoc v2 composite
- r-lib/actions/setup-r v2 composite