conos
R package for the joint analysis of multiple single-cell RNA-seq datasets
Science Score: 23.0%
This score indicates how likely this project is to be science-related based on various indicators:
-
○CITATION.cff file
-
○codemeta.json file
-
○.zenodo.json file
-
✓DOI references
Found 5 DOI reference(s) in README -
○Academic publication links
-
✓Committers with academic emails
2 of 15 committers (13.3%) from academic institutions -
○Institutional organization owner
-
○JOSS paper metadata
-
○Scientific vocabulary similarity
Low similarity (12.8%) to scientific vocabulary
Keywords
Keywords from Contributors
Repository
R package for the joint analysis of multiple single-cell RNA-seq datasets
Basic Info
Statistics
- Stars: 214
- Watchers: 11
- Forks: 39
- Open Issues: 9
- Releases: 0
Topics
Metadata Files
README.md

conos
Conos: Clustering On Network Of Samples
What is conos? Conos is an R package to wire together large collections of single-cell RNA-seq datasets, which allows for both the identification of recurrent cell clusters and the propagation of information between datasets in multi-sample or atlas-scale collections. It focuses on the uniform mapping of homologous cell types across heterogeneous sample collections. For instance, users could investigate a collection of dozens of peripheral blood samples from cancer patients combined with dozens of controls, which perhaps includes samples of a related tissue such as lymph nodes.
How does it work?
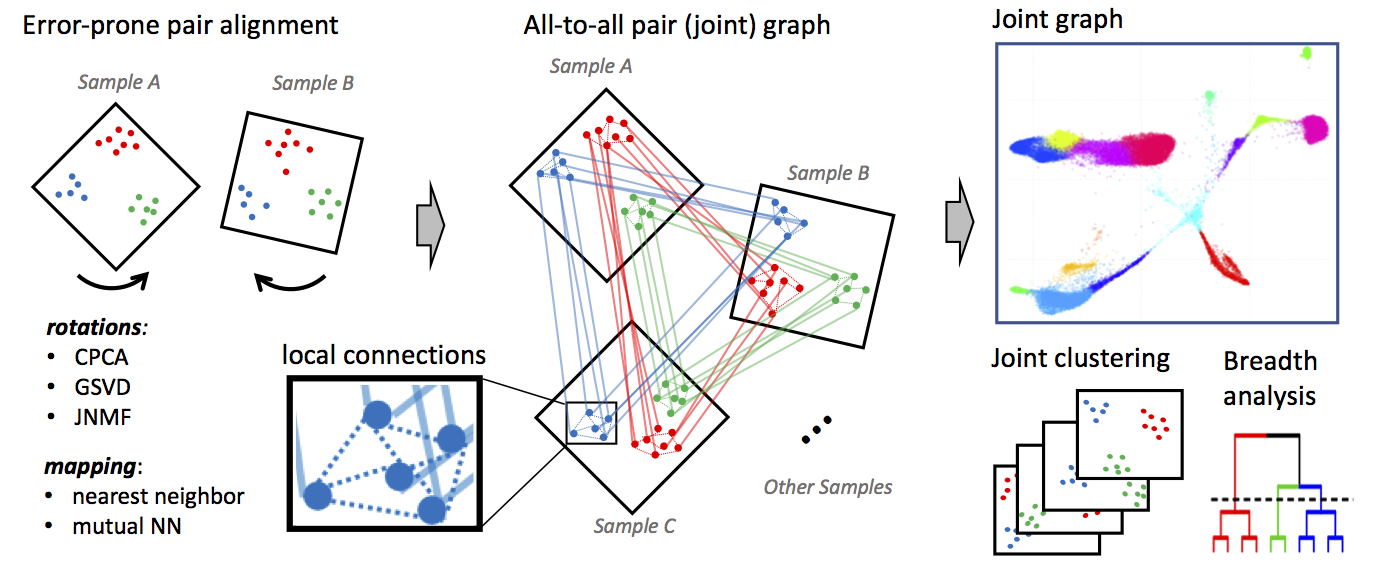 Conos applies one of many error-prone methods to align each pair of samples in a collection, establishing weighted inter-sample cell-to-cell links. The resulting joint graph can then be analyzed to identify subpopulations across different samples. Cells of the same type will tend to map to each other across many such pairwise comparisons, forming cliques that can be recognized as clusters (graph communities).
Conos applies one of many error-prone methods to align each pair of samples in a collection, establishing weighted inter-sample cell-to-cell links. The resulting joint graph can then be analyzed to identify subpopulations across different samples. Cells of the same type will tend to map to each other across many such pairwise comparisons, forming cliques that can be recognized as clusters (graph communities).
Conos processing can be divided into three phases:
* Phase 1: Filtering and normalization Each individual dataset in the sample panel is filtered and normalized using standard packages for single-dataset processing: either pagoda2 or Seurat. Specifically, Conos relies on these methods to perform cell filtering, library size normalization, identification of overdispersed genes and, in the case of pagoda2, variance normalization. (Conos is robust to variations in the normalization procedures, but it is recommended that all of the datasets be processed uniformly.)
* Phase 2: Identify multiple plausible inter-sample mappings Conos performs pairwise comparisons of the datasets in the panel to establish an initial error-prone mapping between cells of different datasets.
* Phase 3: Joint graph construction These inter-sample edges from Phase 2 are then combined with lower-weight intra-sample edges during the joint graph construction. The joint graph is then used for downstream analysis, including community detection and label propagation. For a comprehensive description of the algorithm, please refer to our publication.
What does it produce? In essence, conos will take a large, potentially heterogeneous panel of samples and will produce clustering grouping similar cell subpopulations together in a way that will be robust to inter-sample variation:
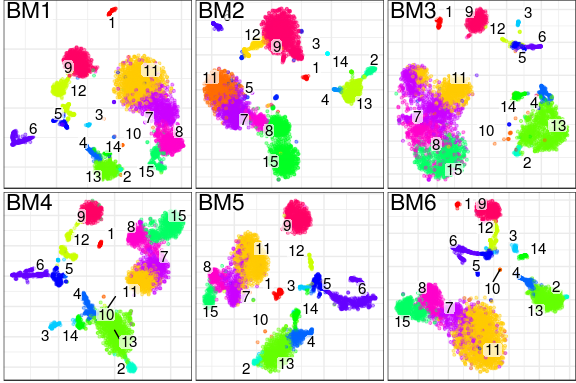
What are the advantages over existing alignment methods? Conos is robust to heterogeneity of samples within a collection, as well as noise. The ability to resolve finer subpopulation structure improves as the size of the panel increases.
Basics of using conos
Given a list of individual processed samples (pl), conos processing can be as simple as this:
```r
Construct Conos object, where pl is a list of pagoda2 objects
con <- Conos$new(pl)
Build joint graph
con$buildGraph()
Find communities
con$findCommunities()
Generate embedding
con$embedGraph()
Plot joint graph
con$plotGraph()
Plot panel with joint clustering results
con$plotPanel() ```
To see more documentation on the class Conos, run ?Conos.
Tutorials
Please see the following tutorials for detailed examples of how to use conos:
Conos walkthrough:
Adjustment of alignment strength with conos:
Integration with Scanpy:
Note that for integration with Scanpy, users need to save conos files to disk from an R session, and then load these files into Python.
Save conos for Scanpy: * HTML version * Markdown version
Load conos files into Scanpy: * Jupyter Notebook
Integrating RNA-seq and ATAC-seq with conos:
Running RNA velocity on a Conos object
First of all, in order to obtain an RNA velocity plot from a Conos object you have to use the dropEst pipeline to align and annotate your single-cell RNA-seq measurements. You can see this tutorial and this shell script to see how it can be done. In this example we specifically assume that when running dropEst you have used the -V option to get estimates of unspliced/spliced counts from the dropEst directly. Secondly, you need the velocyto.R package for the actual velocity estimation and visualisation.
After running dropEst you should have 2 files for each of the samples:
- sample.rds (matrix of counts)
- sample.matrices.rds (3 matrices of exons, introns and spanning reads)
The .matrices.rds files are the velocity files. Load them into R in a list (same order as you give to conos). Load, preprocess and integrate with conos the count matrices (.rds) as you normally would. Before running the velocity, you must at least create an embedding and run the leiden clustering. Finally, you can estimate the velocity as follows:
```r
Assuming con is your Conos object and cms.list is the list of your velocity files
library(velocyto.R)
Preprocess the velocity files to match the Conos object
vi <- velocityInfoConos(cms.list = cms.list, con = con, n.odgenes = 2e3, verbose = TRUE)
Estimate RNA velocity
vel.info <- vi %$% gene.relative.velocity.estimates(emat, nmat, cell.dist = cell.dist, deltaT = 1, kCells = 25, fit.quantile = 0.05, n.cores = 4)
Visualise the velocity on your Conos embedding
Takes a very long time!
Assign to a variable to speed up subsequent recalculations
cc.velo <- show.velocity.on.embedding.cor(vi$emb, vel.info, n = 200, scale = 'sqrt', cell.colors = ac(vi$cell.colors, alpha = 0.5), cex = 0.8, grid.n = 50, cell.border.alpha = 0, arrow.scale = 3, arrow.lwd = 0.6, n.cores = 4, xlab = "UMAP1", ylab = "UMAP2")
Use cc=cc.velo$cc when running again (skips the most time consuming delta projections step)
show.velocity.on.embedding.cor(vi$emb, vel.info, cc = cc.velo$cc, n = 200, scale = 'sqrt', cell.colors = ac(vi$cell.colors, alpha = 0.5), cex = 0.8, arrow.scale = 15, show.grid.flow = TRUE, min.grid.cell.mass = 0.5, grid.n = 40, arrow.lwd = 2, do.par = F, cell.border.alpha = 0.1, n.cores = 4, xlab = "UMAP1", ylab = "UMAP2")
```
Installation
To install the stable version from CRAN, use:
r
install.packages('conos')
To install the latest version of conos, use:
r
install.packages('devtools')
devtools::install_github('kharchenkolab/conos')
System dependencies
The dependencies are inherited from pagoda2. Note that this package also has the dependency igraph, which requires various libraries to install correctly. Please see the installation instructions at that page for more details, along with the github README here.
Ubuntu dependencies
To install system dependencies using apt-get, use the following:
sh
sudo apt-get update
sudo apt-get -y install libcurl4-openssl-dev libssl-dev libxml2-dev libgmp-dev libglpk-dev
Red Hat-based distributions dependencies
For Red Hat distributions using yum, use the following command:
sh
sudo yum update
sudo yum install openssl-devel libcurl-devel libxml2-devel gmp-devel glpk-devel
Mac OS
Using the Mac OS package manager Homebrew, try the following command:
sh
brew update
brew install openssl curl-openssl libxml2 glpk gmp
(You may need to run brew uninstall curl in order for brew install curl-openssl to be successful.)
As of version 1.3.1, conos should successfully install on Mac OS. However, if there are issues, please refer to the following wiki page for further instructions on installing conos with Mac OS: Installing conos for Mac OS
Running conos via Docker
If your system configuration is making it difficult to install conos natively, an alternative way to get conos running is through a docker container.
Note: On Mac OS X, Docker Machine has Memory and CPU limits. To control it, please check instructions either for CLI or for Docker Desktop.
Ready-to-run Docker image
The docker distribution has the latest version and also includes the pagoda2 package. To start a docker container, first install docker on your platform and then start the pagoda2 container with the following command in the shell:
docker run -p 8787:8787 -e PASSWORD=pass pkharchenkolab/conos:latest
The first time you run this command, it will download several large images so make sure that you have fast internet access setup. You can then point your browser to http://localhost:8787/ to get an Rstudio environment with pagoda2 and conos installed (please log in using credentials username=rstudio, password=pass). Explore the docker --mount option to allow access of the docker image to your local files.
Note: If you already downloaded the docker image and want to update it, please pull the latest image with:
docker pull pkharchenkolab/conos:latest
Building Docker image from the Dockerfile
If you want to build image by your own, download the Dockerfile (available in this repo under /docker) and run to following command to build it:
docker build -t conos .
This will create a "conos" docker image on your system (please be patient, as the build could take approximately 30-50 minutes to finish).
You can then run it using the following command:
docker run -d -p 8787:8787 -e PASSWORD=pass --name conos -it conos
References
If you find this software useful for your research, please cite the corresponding paper:
Barkas N., Petukhov V., Nikolaeva D., Lozinsky Y., Demharter S., Khodosevich K., & Kharchenko P.V.
Joint analysis of heterogeneous single-cell RNA-seq dataset collections.
Nature Methods, (2019). doi:10.1038/s41592-019-0466-z
The R package can be cited as:
Viktor Petukhov, Nikolas Barkas, Peter Kharchenko, and Evan
Biederstedt (2021). conos: Clustering on Network of Samples. R
package version 1.5.2.
Owner
- Name: Kharchenko Lab
- Login: kharchenkolab
- Kind: organization
- Website: http://pklab.org
- Twitter: KharchenkoLab
- Repositories: 21
- Profile: https://github.com/kharchenkolab
GitHub Events
Total
- Watch event: 22
- Issue comment event: 4
- Pull request event: 1
- Fork event: 2
Last Year
- Watch event: 22
- Issue comment event: 4
- Pull request event: 1
- Fork event: 2
Committers
Last synced: over 2 years ago
Top Committers
| Name | Commits | |
|---|---|---|
| evanbiederstedt | e****t@g****m | 357 |
| viktor_petukhov | v****v@y****u | 201 |
| Peter Kharchenko | p****o@g****m | 145 |
| Nikolas Barkas | N****s@o****m | 78 |
| evanbiederstedt | e****t | 45 |
| yarloz | l****v@g****m | 43 |
| Nikolas Barkas | N****s@h****u | 24 |
| GMaciag | p****o@g****m | 23 |
| Nikolas Barkas | n****s@o****m | 21 |
| Paul Hoffman | m****e | 13 |
| RRydbirk | r****k@b****k | 3 |
| pkharchenko | p****o@g****m | 3 |
| Darío Hereñú | m****a@g****m | 2 |
| Nikolas Barkas | b****n@N****l | 1 |
| Grzegorz Maciag | 3****g | 1 |
Committer Domains (Top 20 + Academic)
Issues and Pull Requests
Last synced: 7 months ago
All Time
- Total issues: 71
- Total pull requests: 34
- Average time to close issues: 4 months
- Average time to close pull requests: 11 days
- Total issue authors: 41
- Total pull request authors: 5
- Average comments per issue: 4.11
- Average comments per pull request: 2.0
- Merged pull requests: 30
- Bot issues: 0
- Bot pull requests: 0
Past Year
- Issues: 2
- Pull requests: 1
- Average time to close issues: 17 days
- Average time to close pull requests: N/A
- Issue authors: 1
- Pull request authors: 1
- Average comments per issue: 3.5
- Average comments per pull request: 5.0
- Merged pull requests: 0
- Bot issues: 0
- Bot pull requests: 0
Top Authors
Issue Authors
- VPetukhov (12)
- rrydbirk (5)
- ACastanza (5)
- hiraksarkar (3)
- tanasa (3)
- Dimmiso (3)
- akramdi (2)
- GMaciag (2)
- FFriis (2)
- mckoch234 (2)
- bony45 (2)
- auberginekenobi (1)
- tedtoal (1)
- sroyyors (1)
- SaskiaFreytag (1)
Pull Request Authors
- evanbiederstedt (29)
- shivangsharma (2)
- GMaciag (2)
- rrydbirk (1)
- kant (1)
Top Labels
Issue Labels
Pull Request Labels
Packages
- Total packages: 2
-
Total downloads:
- cran 398 last-month
- Total docker downloads: 147
-
Total dependent packages: 2
(may contain duplicates) -
Total dependent repositories: 8
(may contain duplicates) - Total versions: 37
- Total maintainers: 1
proxy.golang.org: github.com/kharchenkolab/conos
- Documentation: https://pkg.go.dev/github.com/kharchenkolab/conos#section-documentation
- License: gpl-3.0
-
Latest release: v1.5.2
published almost 2 years ago
Rankings
cran.r-project.org: conos
Clustering on Network of Samples
- Homepage: https://github.com/kharchenkolab/conos
- Documentation: http://cran.r-project.org/web/packages/conos/conos.pdf
- License: GPL-3
-
Latest release: 1.5.2
published almost 2 years ago
Rankings
Maintainers (1)
Dependencies
- Matrix * depends
- R >= 3.5.0 depends
- igraph * depends
- ComplexHeatmap * imports
- Matrix.utils * imports
- N2R * imports
- R6 * imports
- Rtsne * imports
- abind * imports
- cowplot * imports
- dendextend * imports
- dplyr * imports
- ggplot2 * imports
- ggrepel * imports
- gridExtra * imports
- irlba * imports
- leidenAlg * imports
- magrittr * imports
- methods * imports
- parallel * imports
- reshape2 * imports
- rlang * imports
- sccore >= 1.0.0 imports
- stats * imports
- tools * imports
- utils * imports
- AnnotationDbi * suggests
- BiocParallel * suggests
- DESeq2 * suggests
- GO.db * suggests
- PMA * suggests
- Seurat * suggests
- SummarizedExperiment * suggests
- conosPanel * suggests
- drat * suggests
- entropy * suggests
- ggrastr * suggests
- jsonlite * suggests
- knitr * suggests
- org.Hs.eg.db * suggests
- org.Mm.eg.db * suggests
- p2data * suggests
- pagoda2 * suggests
- plyr * suggests
- rhdf5 * suggests
- rmarkdown * suggests
- rmumps * suggests
- shinycssloaders * suggests
- testthat * suggests
- tibble * suggests
- uwot * suggests
- zoo * suggests