
PyAutoLens
PyAutoLens: Open-Source Strong Gravitational Lensing - Published in JOSS (2021)

Nanomesh
Nanomesh: A Python workflow tool for generating meshes from image data - Published in JOSS (2022)

HistoJS
HistoJS: Web-Based Analytical Tool for Advancing Multiplexed Images - Published in JOSS (2024)

connected-components-3d
Connected components on discrete and continuous multilabel 3D & 2D images. Handles 26, 18, and 6 connected variants; periodic boundaries (4, 8, & 6)


imaging-server-kit
Deploy image processing algorithms in FastAPI servers and easily run them in Napari, QuPath, and more.

symbac
Accurate segmentation of bacterial microscope images using deep learning synthetically generated image data.

regularizepsf
A Python package for manipulating and correcting variable point spread functions.



scenedetect
:movie_camera: Python and OpenCV-based scene cut/transition detection program & library.


catalyst
Accelerated deep learning R&D


simpa
The Simulation and Image Processing for Photonics and Acoustics (SIMPA) toolkit.

mmagic
OpenMMLab Multimodal Advanced, Generative, and Intelligent Creation Toolbox. Unlock the magic 🪄: Generative-AI (AIGC), easy-to-use APIs, awsome model zoo, diffusion models, for text-to-image generation, image/video restoration/enhancement, etc.

frc
frc is a Python package for computing the Fourier Ring Correlation (FRC) of images using DIPlib

pybasic-illumination-correction
Python implementation of the BaSiC shading correction method


mcmicro
An end-to-end processing pipeline that transforms multi-channel whole-slide images into single-cell data.



exllsm-circuit-reconstruction
Nextflow pipeline encapsulating workflows for analyzing expansion lattice light-sheet microscopy (ExLLSM) imagery

edfi
:earth_asia: EDFI is an open-source script to extract data from a 2D or 3D image.


blended-tiling
A seamless / blended tiling module for PyTorch, capable of blending any 4D NCHW tensors together

clustimage
clustimage is a python package for unsupervised clustering of images.

DeconvOptim
A multi-dimensional, high performance deconvolution framework written in Julia Lang for CPUs and GPUs.

spatialvi
Pipeline for processing spatially-resolved gene counts with spatial coordinates and image data. Designed for 10x Genomics Visium transcriptomics.

fft-conv-pytorch
Implementation of 1D, 2D, and 3D FFT convolutions in PyTorch. Much faster than direct convolutions for large kernel sizes.


molkart
A pipeline for processing Molecular Cartography data from Resolve Bioscience (combinatorial FISH)


tf2deepfloorplan
TF2 Deep FloorPlan Recognition using a Multi-task Network with Room-boundary-Guided Attention. Enable tensorboard, quantization, flask, tflite, docker, github actions and google colab.

deeptrack
DeepTrack2 is a modular Python library for generating, manipulating, and analyzing image data pipelines for machine learning and experimental imaging.

sandi
Sediment ANalysis and Delineation through Images. A free and open-source software to process particles from high-resolution underwater images and to analyze gravels from laboratory images. Designed for contour detection, shape characterization and size measurement. SANDI is intended to become a collaborative project.

microaligner
Image registration (alignment) software for large microscopy images

stm-cluster-heightzer
Stm cluster heights resizer (stm-cluster-heightzer) is a class that finds peaks of clusters (high bumps) in a scanning tunneling microscope (STM) image(data) and if necessary corrects the heights of the individual clusters to the surrounding surface.

k-space_wght_msk_for_mri_denoising
k-space weighting and masking for denoising of MRI image without blurring or losing contrast, as well as for brightening of the objects in the image with simultaneous noise reduction (on the example of Agilent FID data). (Python 3)


dosymetric-image-reconstruction
Reconstructing dosimetric images using a deconvoluted physical model


hogpp
Fast computation of rectangular histogram of oriented gradients (R-HOG) features using integral histogram

speckcn2
:satellite: :cyclone: A platform to use speckle patterns to describe atmospheric turbulence

afm-analysis-pipeline
An open-source project that offers an environment in which to construct atomic force microscopy (AFM) image data processing pipelines.


sciview
sciview is a tool for visualization and interaction with ND image and mesh data

thorimage-sync_2p-analysis
Notebook to analyze ThorImage + ThorSync 2-photon data with light stimulation.

mother-db-annotation-tools
This repository contains code for the MOTHER-DB.org specifically related to image segmentation and annotation work flows. MOTHER-DB is a database, meta-data archive and set of programs for annotating and storing ovary histology images from a wide range of species.


mri_k-space-derived_details_edges
k-space based details/edges detection in MRI images with optional k-space based denoising and detail control (on the example of Agilent FID data). (Python 3)
slicer
Multi-platform, free open source software for visualization and image computing.



sgumlp
Pytorch implementation of the SGU-MLP Architecture (mostly) as described in the paper "Spatial Gated Multi-Layer Perceptron for Land Use and Land Cover Mapping".


confocal_z-stack_analysis
Imaging experiments in Biology often involve the acquisition of confocal z-stack images to quantify fluorescent signal in a biological sample. These z-stacks are usually analyzed using open-source programs such as FIJI, which are notoriously frustrating to work with, and do not facility intuitive high-throughput analysis of many images at once. Manual image analysis is labor-intensive, time-consuming, and prone to human error. I compiled two notebooks to streamline two of the most commonly performed tasks in the analysis of confocal z-stack images: the generation of "pretty" maximum projections, and the analysis of pixel values/ fluorescence from sum projections.


deepinv
DeepInverse: a PyTorch library for solving imaging inverse problems using deep learning

fluidimage
:sparkler: A libre Python framework for scientific treatments of large series of images (publish-only mirror)


lsmquant
A pipeline for processing and analysis of light-sheet microscopy images.


complexity-in-complexity
Code for "Complexity in Complexity: Understanding Visual Complexity Through Structure, Color, and Surprise". CogSci, 2025 & ICLR Re-Align Workshop, 2025.

pyhistology
Python package that uses colorspace-based segmentation to analyze histopathology images.

image-crop-analysis
Code for reproducing our analysis in the paper titled: Image Cropping on Twitter: Fairness Metrics, their Limitations, and the Importance of Representation, Design, and Agency

osparc-iseg
The Medical Image Segmentation Tool Set (iSEG) is a fully integrated segmentation (including pre- and postprocessing) toolbox for the efficient, fast, and flexible generation of anatomical models from various types of imaging data


packages_gee
A repository of miscellaneous JavaScript-based packages for Google Earth Engine (GEE)

hyperspectral_toolkit
Google Earth Engine (GEE) JavaScript-based tools for working with various hyperspectral datasets (e.g., AVIRIS, EMIT, Hyperion, PACE OCI)

pace_oci_toolkit
Google Earth Engine (GEE) JavaScript-based tools for working with NASA PACE OCI data


motila
Pipeline for analyzing microglial fine process motility from 4D/5D multi-photon imaging data




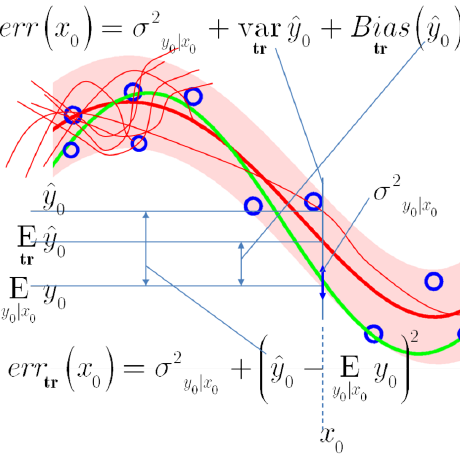
teaching
Teaching Materials for Dr. Waleed A. Yousef

cornea-stretch-and-tonometer-capture-processing
Project Supervised by Professor Jean-Marc Allain on Cornea Stretch Extraction From Tonometer Captures

Colocalization
Colocalization metrics and distances for images or their sparse representations.

exploreasl-gui
A user interface for the analysis of arterial spin labeling (ASL) images. Documentation can be found at: https://mauricepasternak.github.io/ExploreASL-GUI-Docs/latest