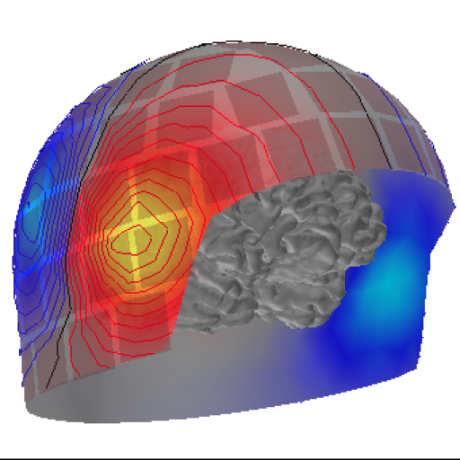
Intracranial Electrode Location and Analysis in MNE-Python
Intracranial Electrode Location and Analysis in MNE-Python - Published in JOSS (2022)

MNE-BIDS
MNE-BIDS: Organizing electrophysiological data into the BIDS format and facilitating their analysis - Published in JOSS (2019)

MNE-ICALabel
MNE-ICALabel: Automatically annotating ICA components with ICLabel in Python - Published in JOSS (2022)

HNN-core
HNN-core: A Python software for cellular and circuit-level interpretation of human MEG/EEG - Published in JOSS (2023)

mTRFpy
mTRFpy: A Python package for temporal response function analysis - Published in JOSS (2023)

SelfEEG
SelfEEG: A Python library for Self-Supervised Learning in Electroencephalography - Published in JOSS (2024)


UnfoldSim.jl
UnfoldSim.jl: Simulating continuous event-based time series data for EEG and beyond - Published in JOSS (2025)

Reducing the efforts to create reproducible analysis code with FieldTrip
Reducing the efforts to create reproducible analysis code with FieldTrip - Published in JOSS (2024)

Connectome Mapper 3
Connectome Mapper 3: A Flexible and Open-Source Pipeline Software for Multiscale Multimodal Human Connectome Mapping - Published in JOSS (2022)

Pycrostates
Pycrostates: a Python library to study EEG microstates - Published in JOSS (2022)

Openseize
Openseize: A digital signal processing package for large EEG datasets in Python - Published in JOSS (2023)

Octopus Sensing
Octopus Sensing: A Python library for human behavior studies - Published in JOSS (2022)

fitgrid
fitgrid: A Python package for multi-channel event-related time series regression modeling - Published in JOSS (2021)

NeuroAnalyzer
NeuroAnalyzer: Julia toolbox for analyzing neurophysiological data - Published in JOSS (2025)



UnfoldMakie
Plotting and visualization tools for EEG data, with additional Unfold.jl regression ERP methods. Based on the visualization libraries Makie.jl and AlgebraOfGraphics.jl

fooof
Parameterizing neural power spectra into periodic & aperiodic components.


pyprep
PyPREP: A Python implementation of the Preprocessing Pipeline (PREP) for EEG data

neurokit2
NeuroKit2: The Python Toolbox for Neurophysiological Signal Processing

pyriemann
Machine learning for multivariate data through the Riemannian geometry of positive definite matrices in Python






brainstorm3
Brainstorm software: MEG, EEG, fNIRS, ECoG, sEEG and electrophysiology



bci
Framework for P300 wave detection and noise-based cyberattacks in Brain-Computer Interfaces - Enrique Tomás Martínez Beltrán







https://github.com/cyclotronresearchcentre/shamo
A tool for electromagnetic modelling of the head and sensitivity analysis.


https://github.com/lokinou/p3k_offline_analysis
P300 speller offline analysis recorded with Openvibe and BCI2000. Using python-MNE


caton-pcb
Hardware design files for wireless EEG system used in concurrent TMS-EEG-fMRI

SPM 25
SPM 25: open source neuroimaging analysis software - Published in JOSS (2025)

eeg-pyline
EEG-pyline is a pipeline for EEG data pre-processing, analysis and visualisation created for neuroscience and mental health research.


eeg_to_erp_pipeline_stats_r
General pipeline used for analyzing EEG data where Raw EEG data gets transformed into ERPS and Stats are done in R (Mixed effects models)

https://github.com/cbrnr/timewarp
Time-warping variable-length EEG epochs for time/frequency analysis

https://github.com/cognitiveneurolab/pipeline_eeg_to_erp_eeglab_stats_r
General pipeline used for analyzing EEG data where Raw EEG data gets transformed into ERPS and Stats are done in R (Mixed effects models)


bids
Template for new EEG studies in NCIL, using BIDS-compliant structure and scripts to import data.

cpp_ptb
a set of function to make it easier to create behavioral, EEG, fMRI experiment with psychtoolbox

areeg-an-open-access-arabic-inner-speech-eeg-dataset
Repository contains all code needed to work with ArEEG dataset


ectmetrics
A Python Library for Calculating Seizure Quality Indices in Electroconvulsive Therapy



oscillationmethods
Methodological considerations for analyzing and interpreting neural oscillations.

developmentaldemo
Tutorial for developmental data with spectral parameterization.

novicevsexpert
Replication package, supplementary materials, and analysis pipeline for our FSE'22 EEG study.


eCobidas
Working repository to turn the COBIDAS guidelines to report methods and results in neuroimaging into a user friendly checklist


jlmerclusterperm
Fast cluster-based permutation test for densely-sampled, multi-level time series data (ex: eyetracking, EEG)



gaitmod
Python library for real-time gait modulation prediction using multimodal neural and movement data (LFP, EEG, IMU, EMG) — designed for closed-loop DBS systems in Parkinson’s disease.

PyRASA - Spectral parametrization in python based on IRASA
PyRASA - Spectral parametrization in python based on IRASA - Published in JOSS (2025)

aoc-frp-code
Replication package, supplementary materials, and analysis pipeline for our EEG study


resting-state-analysis-pipeline-microstates-frequency
Resting state pipeline from pre-processing raw data until doing a power/frequency analysis and a microstate analysis

swat-experiment
Paradigm FKA Swat kids updated. Now contains eyetracking, the ISI is created differently and the the the response window is changed.




https://github.com/braindatalab/calibrain
Python framework for uncertainty estimation and calibration in EEG/MEG inverse source imaging.

https://github.com/john-veillette/niseq
group sequential tests for neuroimaging


eeg-imagined-speech-recognition
Imagined speech recognition using EEG signals. KaraOne database, FEIS database.

eegutils
An R package for processing and plotting of electroencephalography (EEG) data



pyntbci
The Python Noise-Tagging Brain-Computer interface (PyntBCI) library is a Python toolbox for the noise-tagging brain-computer interfacing (BCI) project developed at the Donders Institute for Brain, Cognition and Behaviour, Radboud University, Nijmegen, the Netherlands