
APackOfTheClones
APackOfTheClones: Visualization of clonal expansion with circle packing - Published in JOSS (2024)

Powering single-cell analyses in the browser with WebAssembly
Powering single-cell analyses in the browser with WebAssembly - Published in JOSS (2023)

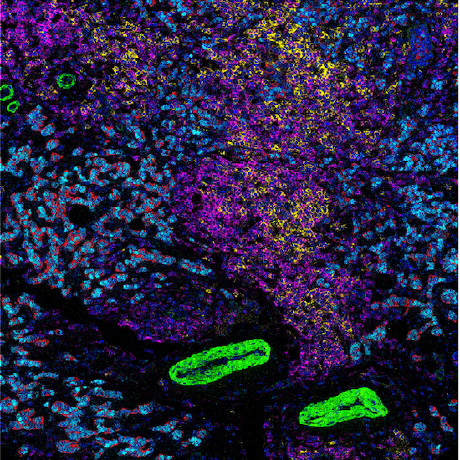
HistoJS
HistoJS: Web-Based Analytical Tool for Advancing Multiplexed Images - Published in JOSS (2024)

vitessce
Vitessce is a visual integration tool for exploration of spatial single-cell experiments.

scrnaseq
Single-cell RNA-Seq pipeline for barcode-based protocols such as 10x, DropSeq or SmartSeq, offering a variety of aligners and empty-droplet detection


celltypist
A tool for semi-automatic cell type classification

cytodataframe
An in-memory data analysis format for single-cell profiles alongside their corresponding images and segmentation masks.


BASiCS
BASiCS: Bayesian Analysis of Single-Cell Sequencing Data. This is an unstable experimental version. Please see http://bioconductor.org/packages/BASiCS/ for the official release version



mcmicro
An end-to-end processing pipeline that transforms multi-channel whole-slide images into single-cell data.

mllmcelltype
🏆 #1 Multi-LLM consensus framework | 550+ stars | 95% accuracy | 10+ LLM providers | Leading cell annotation tool



mixscape_seurat
A Snakemake workflow and MrBiomics module for performing perturbation analyses of pooled (multimodal) CRISPR screens with sc/snRNA-seq read-out (scCRISPR-seq) powered by the R package Seurat's method Mixscape.

dea_seurat
A Snakemake workflow and MrBiomics module for performing differential expression analyses (DEA) on (multimodal) sc/snRNA-seq data powered by the R package Seurat.

pixelgen-pixelator
A command-line tool and library to process and analyze sequencing data from Molecular Pixelation (MPX) and Proximity Network (PNA) assays.

trisicell
Scalable tumor phylogeny inference and validation from single-cell RNA or DNA data

cna
Covarying neighborhood analysis (CNA) is a method for finding structure in- and conducting association analysis with multi-sample single-cell datasets.


single-cell-training
SIB course on single cell transcriptomics by mostly using the Seurat pipeline


sccomp
Mixed-effect model to test differences in cell type proportions from single-cell data, in R

sccustomize
R package with collection of functions created and/or curated to aid in the visualization and analysis of single-cell data using R.

spatialvi
Pipeline for processing spatially-resolved gene counts with spatial coordinates and image data. Designed for 10x Genomics Visium transcriptomics.


scpubr
Generate high quality, publication ready visualizations for single cell transcriptomics data.


molkart
A pipeline for processing Molecular Cartography data from Resolve Bioscience (combinatorial FISH)


scperturb
scPerturb: A resource and a python/R tool for single-cell perturbation data


iSEE
R/shiny interface for interactive visualization of data in SummarizedExperiment objects
gcmodeller
GCModeller: genomics CAD(Computer Assistant Design) Modeller system in .NET language



EWCE
Expression Weighted Celltype Enrichment. See the package website for up-to-date instructions on usage.

singlecell-qtl
Discovery and characterization of variance QTLs in human induced pluripotent stem cells

pagoda2
R package for analyzing and interactively exploring large-scale single-cell RNA-seq datasets



netSmooth
netSmooth: A Network smoothing based method for Single Cell RNA-seq imputation

scCATCH
Automatic Annotation on Cell Types of Clusters from Single-Cell RNA Sequencing Data

cite-seq-count
A tool that allows to get UMI counts from a single cell protein assay

pyscenic
pySCENIC is a lightning-fast python implementation of the SCENIC pipeline (Single-Cell rEgulatory Network Inference and Clustering) which enables biologists to infer transcription factors, gene regulatory networks and cell types from single-cell RNA-seq data.

itclust
Iterative transfer learning with neural network improves clustering and cell type classification in single-cell RNA-seq analysis

https://github.com/cbg-ethz/scicone
Single-cell copy number calling and event history reconstruction.

https://github.com/becavin-lab/checkatlas
One liner tool to check the quality of your single-cell atlases.

scMerge
Statistical approach for removing unwanted variation from multiple single-cell datasets

escheR
R package to visualize colocalization for single cell & spatially-resolved genomics data

AnanseSeurat
Single cell ANANSE Gene-regulatory-network analysis from Seurat objects




digitaldlsorter
digitalDLSorteR: An R package to deconvolute bulk RNA-Seq using scRNA-Seq data

disc
A highly scalable and accurate inference of gene expression and structure for single-cell transcriptomes using semi-supervised deep learning.

cellama
Cell type annotation with local Large Language Models (LLMs) - Ensuring privacy and speed with extensive customized reports



nebulosa
R package to visualize gene expression data based on weighted kernel density estimation

https://github.com/bioconductor/orchestratingsinglecellanalysis
Content for the OSCA Book.

scrnaseq_processing_seurat
A Snakemake workflow and MrBiomics module for processing and visualizing (multimodal) sc/snRNA-seq data generated with 10X Genomics Kits or in the MTX matrix file format powered by the R package Seurat.


pixelator
Pipeline to generate Molecular Pixelation data with Pixelator (Pixelgen Technologies AB)

easybio
Comprehensive Single-Cell Annotation and Transcriptomic Analysis Toolkit

ccsinglecell
A project focused on using single-cell RNA sequencing data (scRNA-seq) and pseudo time to improve colon cancer diagnosis and outcomes.

https://github.com/cellgeni/nf-atac
Pipeline to process sc-ATAC data with pyCistopic


neuroblastoma
Single-cell transcriptomics and epigenomics unravel the role of monocytes in neuroblastoma bone marrow metastasis


sdevelo
Multivariate stochastic modeling for transcriptional dynamics with cell-specific latent time using SDEvelo

scdownstream
A single cell transcriptomics pipeline for QC, integration and making the data presentable




https://github.com/alexslemonade/scpca-portal
Single-cell Pediatric Cancer Atlas Portal is a growing database of uniformly processed single-cell data from pediatric cancer tumors and model systems


mzkit
Data toolkits for processing NMR, MALDI MSI, MALDI single cell, Raman Spectroscopy, LC-MS and GC-MS raw data, chemoinformatics data analysis and data visualization.

pyclustree
A Python alternative to `clustree` for assessing single-cell RNA-sequencing clusters.

https://github.com/cadaei-yuvxvs/snrnaseq_muscle_mnd
Single Nuclei RNA Sequencing analysis of MND muscle cultures for the paper "Impaired signaling for neuromuscular synaptic maintenance is a feature of Motor Neuron Disease" by Ding et al, published in Acta Neuropathologica Communications.


https://github.com/alexslemonade/openscpca-analysis
An open, collaborative project to analyze data from the Single-cell Pediatric Cancer Atlas (ScPCA) Portal

chemcpa
Code for "Predicting Cellular Responses to Novel Drug Perturbations at a Single-Cell Resolution", NeurIPS 2022.

fsctype
Modification of the Original ScType Method for Barcode Annotation using KNN Algorithm

scmorph
scmorph: a Python package for analysing single-cell morphological profiles - Published in JOSS (2025)

tidytof
An R package for analyzing high-dimensional cytometry data using the tidyverse.

scgate
marker-based purification of cell types from single-cell RNA-seq datasets

scnanoseq
Single-cell/nuclei pipeline for data derived from Oxford Nanopore and 10X Genomics


https://github.com/cafferychen777/awesome-scrna-annotation
A curated list of tools and methods for scRNA-seq cell type annotation

https://github.com/bvieth/powsimr
Power analysis is essential to optimize the design of RNA-seq experiments and to assess and compare the power to detect differentially expressed genes. PowsimR is a flexible tool to simulate and evaluate differential expression from bulk and especially single-cell RNA-seq data making it suitable for a priori and posterior power analyses.

https://github.com/bodenmillergroup/imcdataanalysis
R based workflow for multiplexed imaging data

singleCellHaystack
Finding surprising needles (=genes) in haystacks (=single cell transcriptome data).

fucci-seq
Characterizing and inferring quantitative cell cycle phase in single-cell RNA-seq data analysis